
| Name: LOC100289365 | Sequence: fasta or formatted (295aa) | NCBI GI: 239743009 | |
|
Description: PREDICTED: hypothetical protein XP_002342695
|
Referenced in: DNA Transposon and Retrovirus-related Sequences
| ||
|
Composition:
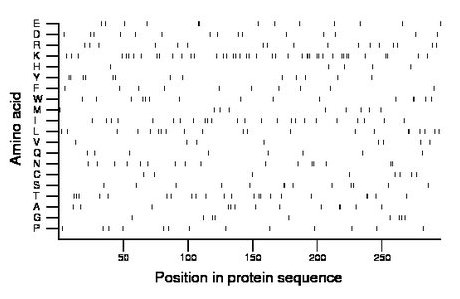
Amino acid Percentage Count Longest homopolymer A alanine 4.4 13 2 C cysteine 2.7 8 1 D aspartate 5.1 15 1 E glutamate 4.4 13 2 F phenylalanine 3.4 10 1 G glycine 3.1 9 1 H histidine 1.7 5 1 I isoleucine 8.1 24 1 K lysine 14.6 43 3 L leucine 7.1 21 2 M methionine 3.1 9 1 N asparagine 5.4 16 2 P proline 4.4 13 1 Q glutamine 2.7 8 1 R arginine 5.8 17 1 S serine 4.4 13 2 T threonine 7.5 22 2 V valine 2.7 8 1 W tryptophan 5.1 15 2 Y tyrosine 4.4 13 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342695 LOC100293629 1.000 PREDICTED: similar to putative p150 LOC100291256 1.000 PREDICTED: hypothetical protein XP_002346845 LOC100130764 0.616 PREDICTED: p150-like LOC100130764 0.616 PREDICTED: p150-like LOC100130764 0.612 PREDICTED: p150-like FLJ10088 0.494 PREDICTED: hypothetical protein FLJ10088 0.494 PREDICTED: hypothetical protein FLJ10088 0.494 PREDICTED: hypothetical protein LOC100292074 0.369 PREDICTED: hypothetical protein XP_002345940 LOC100290874 0.369 PREDICTED: hypothetical protein XP_002346807 LOC100294314 0.369 PREDICTED: hypothetical protein XP_002344084 LOC100294273 0.369 PREDICTED: hypothetical protein XP_002344051 LOC100290606 0.259 PREDICTED: hypothetical protein XP_002347335 LOC100291812 0.192 PREDICTED: hypothetical protein XP_002344872 LOC100290636 0.192 PREDICTED: hypothetical protein XP_002347566 LOC100289563 0.192 PREDICTED: hypothetical protein XP_002343386 TNRC6B 0.010 trinucleotide repeat containing 6B isoform 2 TNRC6B 0.010 trinucleotide repeat containing 6B isoform 1 C7orf31 0.010 hypothetical protein LOC136895 KIAA0586 0.008 talpid3 protein SR140 0.008 U2-associated SR140 protein SPESP1 0.007 sperm equatorial segment protein 1 RAB12 0.007 RAB12, member RAS oncogene family NOC3L 0.007 nucleolar complex associated 3 homolog ADRBK1 0.007 beta-adrenergic receptor kinase 1 DNAH5 0.007 dynein, axonemal, heavy chain 5 DYSF 0.005 dysferlin isoform 2 DYSF 0.005 dysferlin isoform 8 DYSF 0.005 dysferlin isoform 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
