
| Name: LOC100134102 | Sequence: fasta or formatted (86aa) | NCBI GI: 169194762 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
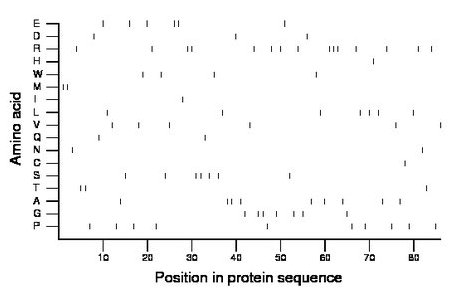
Amino acid Percentage Count Longest homopolymer A alanine 10.5 9 2 C cysteine 1.2 1 1 D aspartate 3.5 3 1 E glutamate 7.0 6 2 F phenylalanine 0.0 0 0 G glycine 8.1 7 2 H histidine 1.2 1 1 I isoleucine 1.2 1 1 K lysine 0.0 0 0 L leucine 8.1 7 1 M methionine 2.3 2 2 N asparagine 2.3 2 1 P proline 11.6 10 1 Q glutamine 2.3 2 1 R arginine 17.4 15 3 S serine 8.1 7 2 T threonine 3.5 3 2 V valine 7.0 6 1 W tryptophan 4.7 4 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein FAM21C 0.420 hypothetical protein LOC253725 FAM21A 0.414 hypothetical protein LOC387680 HNRPDL 0.032 heterogeneous nuclear ribonucleoprotein D-like [Homo... LOC100292294 0.032 PREDICTED: hypothetical protein LOC100289924 0.032 PREDICTED: hypothetical protein XP_002346722 LOC100287776 0.032 PREDICTED: hypothetical protein XP_002342571 LOC100293167 0.025 PREDICTED: hypothetical protein LOC100290803 0.025 PREDICTED: hypothetical protein XP_002347798 LOC100287250 0.025 PREDICTED: hypothetical protein XP_002344493 LOC100287525 0.025 PREDICTED: hypothetical protein XP_002343590 BTBD17 0.025 BTB (POZ) domain containing 17 UNC84A 0.019 unc-84 homolog A isoform a UNC84A 0.019 unc-84 homolog A isoform b MEPCE 0.019 bin3, bicoid-interacting 3 PRIC285 0.019 PPAR-alpha interacting complex protein 285 isoform ... LOC100291617 0.019 PREDICTED: hypothetical protein XP_002344785 LOC100286897 0.019 PREDICTED: hypothetical protein LOC100286897 0.019 PREDICTED: hypothetical protein XP_002344122 RBM15 0.013 RNA binding motif protein 15 LOC100287820 0.013 PREDICTED: hypothetical protein XP_002343297 C15orf52 0.013 hypothetical protein LOC388115 LOC100291727 0.013 PREDICTED: hypothetical protein XP_002345242 LOC100290083 0.013 PREDICTED: hypothetical protein XP_002348010 LOC100289217 0.013 PREDICTED: hypothetical protein XP_002343686 ITPR2 0.013 inositol 1,4,5-triphosphate receptor, type 2 LOC100292296 0.006 PREDICTED: hypothetical protein LOC100290476 0.006 PREDICTED: hypothetical protein XP_002347412 LOC100127891 0.006 PREDICTED: similar to hCG2042508 LOC100127891 0.006 PREDICTED: similar to hCG2042508Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
