
| Name: TOMM22 | Sequence: fasta or formatted (142aa) | NCBI GI: 9910382 | |
|
Description: mitochondrial import receptor Tom22
|
Referenced in:
| ||
|
Composition:
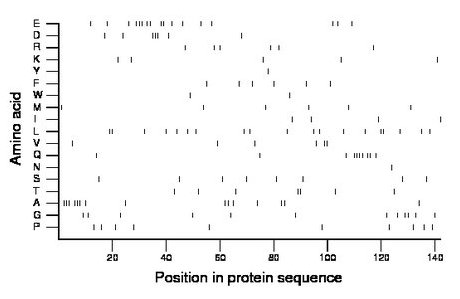
Amino acid Percentage Count Longest homopolymer A alanine 10.6 15 3 C cysteine 0.0 0 0 D aspartate 4.9 7 3 E glutamate 12.0 17 3 F phenylalanine 4.2 6 1 G glycine 8.5 12 2 H histidine 0.0 0 0 I isoleucine 2.8 4 1 K lysine 3.5 5 1 L leucine 13.4 19 2 M methionine 4.2 6 1 N asparagine 0.7 1 1 P proline 7.0 10 1 Q glutamine 7.0 10 4 R arginine 4.2 6 1 S serine 5.6 8 1 T threonine 4.9 7 2 V valine 4.2 6 2 W tryptophan 1.4 2 1 Y tyrosine 0.7 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 mitochondrial import receptor Tom22 CASZ1 0.043 castor homolog 1, zinc finger isoform a ARX 0.043 aristaless related homeobox DEK 0.035 DEK oncogene isoform 1 DEK 0.035 DEK oncogene isoform 2 AEBP2 0.031 AE binding protein 2 isoform b HOMEZ 0.031 homeodomain leucine zipper protein AEBP2 0.031 AE binding protein 2 isoform a SCAF1 0.027 SR-related CTD-associated factor 1 BCCIP 0.027 BRCA2 and CDKN1A-interacting protein isoform C [Homo... BCCIP 0.027 BRCA2 and CDKN1A-interacting protein isoform BCCIPbe... BCCIP 0.027 BRCA2 and CDKN1A-interacting protein isoform BCCIPalp... WDR78 0.027 WD repeat domain 78 isoform 1 WDR78 0.027 WD repeat domain 78 isoform 2 SLC9A10 0.023 sperm-specific sodium proton exchanger TAF7L 0.023 TATA box binding protein-associated factor, RNA poly... LOC100291923 0.023 PREDICTED: hypothetical protein XP_002344991 LOC100291342 0.023 PREDICTED: hypothetical protein XP_002347636 LOC100287346 0.023 PREDICTED: hypothetical protein XP_002343486 ROBO4 0.023 roundabout homolog 4, magic roundabout MAP1B 0.023 microtubule-associated protein 1B EIF5B 0.023 eukaryotic translation initiation factor 5B ANO8 0.020 anoctamin 8 FAM123B 0.020 family with sequence similarity 123B SALL2 0.020 sal-like 2 MYLK 0.020 myosin light chain kinase isoform 3A MYLK 0.020 myosin light chain kinase isoform 1 MYLK 0.020 myosin light chain kinase isoform 3B MYLK 0.020 myosin light chain kinase isoform 2 NOL3 0.020 nucleolar protein 3Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
