
| Name: AMTN | Sequence: fasta or formatted (209aa) | NCBI GI: 47086439 | |
|
Description: amelotin
|
Referenced in:
| ||
|
Composition:
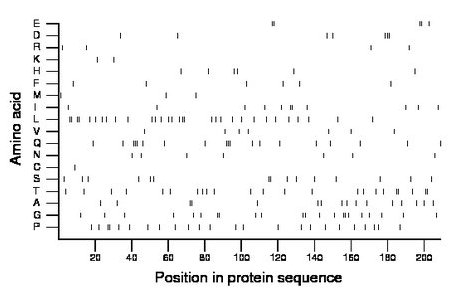
Amino acid Percentage Count Longest homopolymer A alanine 7.7 16 2 C cysteine 0.5 1 1 D aspartate 3.3 7 3 E glutamate 2.4 5 2 F phenylalanine 2.9 6 1 G glycine 10.5 22 2 H histidine 2.9 6 1 I isoleucine 5.3 11 2 K lysine 1.0 2 1 L leucine 14.8 31 2 M methionine 1.4 3 1 N asparagine 3.3 7 1 P proline 11.5 24 2 Q glutamine 9.1 19 3 R arginine 1.9 4 1 S serine 7.2 15 2 T threonine 11.0 23 2 V valine 3.3 7 1 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 amelotin OTOG 0.030 PREDICTED: otogelin isoform 1 OTOG 0.030 PREDICTED: otogelin isoform 2 OTOG 0.030 PREDICTED: otogelin isoform 2 OTOG 0.030 PREDICTED: otogelin isoform 1 OTOG 0.030 PREDICTED: otogelin isoform 2 OTOG 0.030 PREDICTED: otogelin isoform 1 HGC6.3 0.025 hypothetical protein LOC100128124 KIAA1618 0.023 hypothetical protein LOC57714 GORASP2 0.020 golgi reassembly stacking protein 2 ADNP2 0.020 ADNP homeobox 2 CRKRS 0.020 Cdc2-related kinase, arginine/serine-rich isoform 2... CRKRS 0.020 Cdc2-related kinase, arginine/serine-rich isoform 1... MED25 0.020 mediator complex subunit 25 SYN1 0.018 synapsin I isoform Ia SYN1 0.018 synapsin I isoform Ib LOC100292392 0.015 PREDICTED: hypothetical protein LOC647055 0.015 PREDICTED: similar to hCG1790759 LOC100287377 0.015 PREDICTED: hypothetical protein LOC647055 0.015 PREDICTED: similar to hCG1790759 LOC100287377 0.015 PREDICTED: hypothetical protein XP_002342514 NUMBL 0.015 numb homolog (Drosophila)-like ZBTB45 0.015 zinc finger and BTB domain containing 45 CDC2L5 0.013 cell division cycle 2-like 5 isoform 1 CPSF6 0.013 cleavage and polyadenylation specific factor 6, 68 ... CAMKV 0.013 CaM kinase-like vesicle-associated DDAH2 0.013 dimethylarginine dimethylaminohydrolase 2 GLTSCR1 0.013 glioma tumor suppressor candidate region gene 1 [Ho... RBM12 0.013 RNA binding motif protein 12 RBM12 0.013 RNA binding motif protein 12Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
