
| Name: NUP210 | Sequence: fasta or formatted (1887aa) | NCBI GI: 27477134 | |
|
Description: nucleoporin 210
|
Referenced in:
| ||
|
Composition:
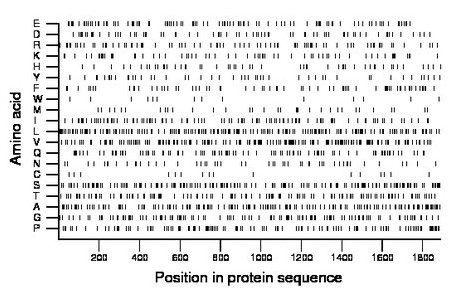
Amino acid Percentage Count Longest homopolymer A alanine 8.5 160 4 C cysteine 0.8 16 1 D aspartate 3.8 71 2 E glutamate 5.2 98 2 F phenylalanine 3.8 72 2 G glycine 6.3 119 2 H histidine 2.9 54 2 I isoleucine 5.6 105 2 K lysine 3.4 65 2 L leucine 10.1 191 4 M methionine 1.8 34 1 N asparagine 2.8 52 2 P proline 6.5 122 2 Q glutamine 5.1 96 2 R arginine 4.6 86 2 S serine 9.1 172 4 T threonine 6.5 123 2 V valine 10.0 188 3 W tryptophan 0.7 14 1 Y tyrosine 2.6 49 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 nucleoporin 210 NUP210L 0.414 nucleoporin 210kDa-like isoform 1 NUP210L 0.391 nucleoporin 210kDa-like isoform 2 LOC100292626 0.034 PREDICTED: similar to Putative uncharacterized prot... LOC100292874 0.014 PREDICTED: similar to hCG1641141 LOC100288782 0.014 PREDICTED: hypothetical protein XP_002342363 SLFN12 0.003 schlafen family member 12 OTC 0.003 ornithine carbamoyltransferase precursor SDC1 0.002 syndecan 1 precursor SDC1 0.002 syndecan 1 precursor DSCAML1 0.002 Down syndrome cell adhesion molecule like 1 KIAA0754 0.002 hypothetical protein LOC643314 TMPRSS13 0.002 transmembrane protease, serine 13 MARCH5 0.002 membrane-associated ring finger (C3HC4) 5 ARHGAP23 0.002 Rho GTPase activating protein 23 MUC16 0.002 mucin 16 NEK4 0.002 NIMA-related kinase 4 FAM83C 0.002 hypothetical protein LOC128876 FOXD2 0.002 forkhead box D2 BCL9L 0.002 B-cell CLL/lymphoma 9-likeHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
