
| Name: LOC100291845 | Sequence: fasta or formatted (139aa) | NCBI GI: 239756990 | |
|
Description: PREDICTED: hypothetical protein XP_002345132
| Not currently referenced in the text | ||
|
Composition:
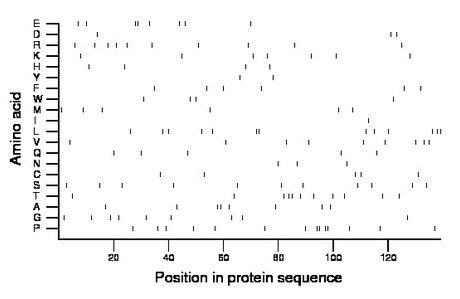
Amino acid Percentage Count Longest homopolymer A alanine 5.8 8 2 C cysteine 3.6 5 1 D aspartate 2.2 3 1 E glutamate 5.8 8 2 F phenylalanine 4.3 6 1 G glycine 6.5 9 1 H histidine 2.9 4 1 I isoleucine 0.7 1 1 K lysine 5.0 7 1 L leucine 9.4 13 2 M methionine 4.3 6 1 N asparagine 2.2 3 1 P proline 10.1 14 2 Q glutamine 3.6 5 1 R arginine 7.2 10 1 S serine 7.9 11 1 T threonine 7.9 11 2 V valine 6.5 9 1 W tryptophan 2.9 4 1 Y tyrosine 1.4 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002345132 LOC100290794 0.985 PREDICTED: hypothetical protein XP_002347877 LOC100288839 0.985 PREDICTED: hypothetical protein XP_002344178 CELSR3 0.029 cadherin EGF LAG seven-pass G-type receptor 3 [Homo... LOC100293445 0.018 PREDICTED: hypothetical protein LOC100288225 0.018 PREDICTED: hypothetical protein LOC100288225 0.018 PREDICTED: hypothetical protein XP_002343333 SIM2 0.015 single-minded homolog 2 long isoform SIM2 0.015 single-minded homolog 2 short isoform FUT3 0.011 fucosyltransferase 3 FUT3 0.011 fucosyltransferase 3 MUC2 0.011 mucin 2 precursor FUT3 0.011 fucosyltransferase 3 FUT3 0.011 fucosyltransferase 3 TACC2 0.011 transforming, acidic coiled-coil containing protein... TACC2 0.011 transforming, acidic coiled-coil containing protein ... TACC2 0.011 transforming, acidic coiled-coil containing protein ... TACC2 0.011 transforming, acidic coiled-coil containing protein ... C6orf136 0.007 hypothetical protein LOC221545 isoform 3 C6orf136 0.007 hypothetical protein LOC221545 isoform 1 LOC100130298 0.007 PREDICTED: hypothetical protein LOC100130298 LOC100130298 0.007 PREDICTED: hypothetical protein LOC100130298 LOC100130298 0.007 PREDICTED: hypothetical protein LOC100130298 AUTS2 0.007 autism susceptibility candidate 2 isoform 2 AUTS2 0.007 autism susceptibility candidate 2 isoform 1 INPP5E 0.007 inositol polyphosphate-5-phosphatase E FUT6 0.007 fucosyltransferase 6 FUT6 0.007 fucosyltransferase 6 BAI3 0.007 brain-specific angiogenesis inhibitor 3 CHD3 0.007 chromodomain helicase DNA binding protein 3 isoform ...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
