
| Name: LOC100132767 | Sequence: fasta or formatted (99aa) | NCBI GI: 169210203 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {99aa} PREDICTED: hypothetical protein | |||
|
Composition:
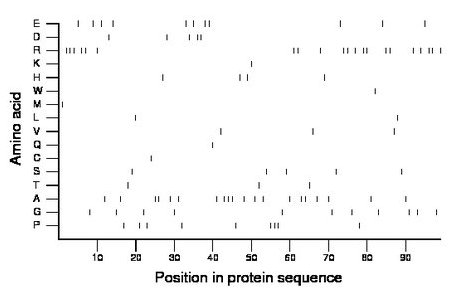
Amino acid Percentage Count Longest homopolymer A alanine 20.2 20 3 C cysteine 1.0 1 1 D aspartate 5.1 5 2 E glutamate 11.1 11 2 F phenylalanine 0.0 0 0 G glycine 11.1 11 1 H histidine 4.0 4 1 I isoleucine 0.0 0 0 K lysine 1.0 1 1 L leucine 2.0 2 1 M methionine 1.0 1 1 N asparagine 0.0 0 0 P proline 9.1 9 3 Q glutamine 1.0 1 1 R arginine 21.2 21 3 S serine 5.1 5 1 T threonine 3.0 3 1 V valine 3.0 3 1 W tryptophan 1.0 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100132767 1.000 PREDICTED: hypothetical protein LOC730060 0.633 PREDICTED: hypothetical protein LOC730060 0.633 PREDICTED: hypothetical protein LOC730060 0.633 PREDICTED: hypothetical protein DOT1L 0.061 DOT1-like, histone H3 methyltransferase IRF7 0.056 interferon regulatory factor 7 isoform b LMTK3 0.056 lemur tyrosine kinase 3 FLJ37078 0.050 hypothetical protein LOC222183 SLC4A3 0.044 solute carrier family 4, anion exchanger, member 3 ... SLC4A3 0.044 solute carrier family 4, anion exchanger, member 3 ... FBRSL1 0.044 fibrosin-like 1 GPR153 0.044 G protein-coupled receptor 153 LOC100292328 0.039 PREDICTED: hypothetical protein LOC100292934 0.039 PREDICTED: hypothetical protein LOC729240 0.039 PREDICTED: FLJ40296 protein family member LOC100291243 0.039 PREDICTED: hypothetical protein XP_002348077 LOC100287025 0.039 PREDICTED: hypothetical protein LOC100287025 0.039 PREDICTED: hypothetical protein XP_002344222 MCF2L 0.039 MCF.2 cell line derived transforming sequence-like ... MCF2L 0.039 MCF.2 cell line derived transforming sequence-like ... PRR20 0.039 proline rich 20 ZNF541 0.039 zinc finger protein 541 LOC729250 0.039 hypothetical protein LOC729250 LOC729246 0.039 hypothetical protein LOC729246 LOC729240 0.039 hypothetical protein LOC729240 LOC729233 0.039 hypothetical protein LOC729233 MEPCE 0.039 bin3, bicoid-interacting 3 LOC100127930 0.039 PREDICTED: hypothetical protein LOC100127930 0.039 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
