
| Name: HDGFL1 | Sequence: fasta or formatted (251aa) | NCBI GI: 110681712 | |
|
Description: hepatoma derived growth factor-like 1
|
Referenced in:
| ||
|
Composition:
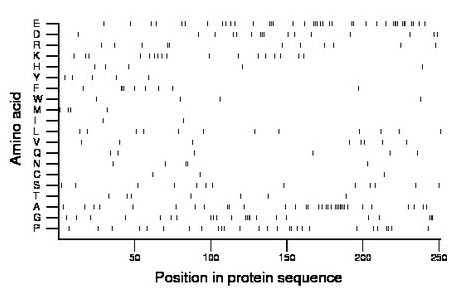
Amino acid Percentage Count Longest homopolymer A alanine 13.5 34 7 C cysteine 1.2 3 1 D aspartate 6.4 16 3 E glutamate 15.9 40 4 F phenylalanine 3.6 9 3 G glycine 9.2 23 4 H histidine 2.0 5 1 I isoleucine 0.8 2 1 K lysine 6.8 17 1 L leucine 4.8 12 1 M methionine 1.6 4 1 N asparagine 2.0 5 2 P proline 10.8 27 2 Q glutamine 2.4 6 1 R arginine 4.4 11 2 S serine 5.2 13 1 T threonine 2.8 7 1 V valine 3.2 8 1 W tryptophan 1.6 4 1 Y tyrosine 2.0 5 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hepatoma derived growth factor-like 1 HDGF 0.447 hepatoma-derived growth factor isoform a HDGF 0.375 hepatoma-derived growth factor isoform c HDGF 0.375 hepatoma-derived growth factor isoform b HDGF2 0.244 hepatoma-derived growth factor-related protein 2 iso... HDGF2 0.244 hepatoma-derived growth factor-related protein 2 iso... HDGFRP3 0.224 hepatoma-derived growth factor, related protein 3 [Ho... PSIP1 0.220 PC4 and SFRS1 interacting protein 1 isoform 2 [Homo... PSIP1 0.220 PC4 and SFRS1 interacting protein 1 isoform 1 [Homo... PSIP1 0.220 PC4 and SFRS1 interacting protein 1 isoform 2 CLIC6 0.054 chloride intracellular channel 6 N-PAC 0.050 cytokine-like nuclear factor n-pac OGFR 0.040 opioid growth factor receptor SIRT1 0.040 sirtuin 1 isoform a HNRNPUL2 0.040 heterogeneous nuclear ribonucleoprotein U-like 2 [H... MDN1 0.040 MDN1, midasin homolog KIAA0754 0.038 hypothetical protein LOC643314 BASP1 0.038 brain abundant, membrane attached signal protein 1 [... RP1L1 0.038 retinitis pigmentosa 1-like 1 FAM9A 0.038 family with sequence similarity 9, member A MARCKS 0.036 myristoylated alanine-rich protein kinase C substra... PPP1R15A 0.036 protein phosphatase 1, regulatory subunit 15A [Homo... NEFH 0.036 neurofilament, heavy polypeptide 200kDa MYST4 0.036 MYST histone acetyltransferase (monocytic leukemia)... NCL 0.034 nucleolin DNMT3A 0.034 DNA cytosine methyltransferase 3 alpha isoform a [Ho... DNMT3A 0.034 DNA cytosine methyltransferase 3 alpha isoform a [Ho... DNMT3A 0.034 DNA cytosine methyltransferase 3 alpha isoform b [Ho... RCSD1 0.034 RCSD domain containing 1 SLC38A10 0.032 solute carrier family 38, member 10 isoform aHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
