
| Name: LOC100292724 | Sequence: fasta or formatted (127aa) | NCBI GI: 239758079 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
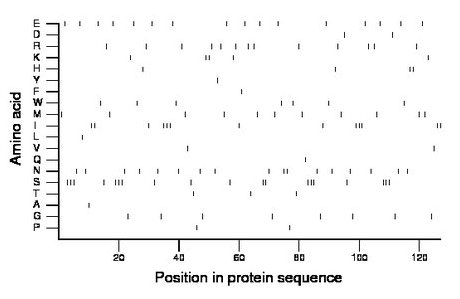
Amino acid Percentage Count Longest homopolymer A alanine 0.8 1 1 C cysteine 0.0 0 0 D aspartate 1.6 2 1 E glutamate 12.6 16 1 F phenylalanine 0.8 1 1 G glycine 6.3 8 1 H histidine 3.1 4 2 I isoleucine 10.2 13 3 K lysine 3.9 5 2 L leucine 0.8 1 1 M methionine 8.7 11 1 N asparagine 13.4 17 2 P proline 1.6 2 1 Q glutamine 0.8 1 1 R arginine 10.2 13 1 S serine 15.0 19 3 T threonine 2.4 3 1 V valine 1.6 2 1 W tryptophan 5.5 7 1 Y tyrosine 0.8 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291599 0.898 PREDICTED: hypothetical protein XP_002346265 LOC100291569 0.787 PREDICTED: hypothetical protein XP_002346264 LOC100293418 0.758 PREDICTED: hypothetical protein LOC100293021 0.529 PREDICTED: hypothetical protein LOC100293793 0.410 PREDICTED: hypothetical protein LOC100293129 0.176 PREDICTED: hypothetical protein LOC642424 0.123 PREDICTED: similar to hCG1742442 SMARCAL1 0.016 SWI/SNF-related matrix-associated actin-dependent re... SMARCAL1 0.016 SWI/SNF-related matrix-associated actin-dependent r... IPMK 0.012 inositol polyphosphate multikinase PAPOLG 0.008 poly(A) polymerase gamma ESF1 0.008 ABT1-associated protein CCDC66 0.004 coiled-coil domain containing 66 isoform 1 CCDC66 0.004 coiled-coil domain containing 66 isoform 2 NLRP12 0.004 NLR family, pyrin domain containing 12 isoform 2 [Ho... LOC100294457 0.004 PREDICTED: similar to hCG2038988 SAV1 0.004 WW45 protein AMY2A 0.004 pancreatic amylase alpha 2A precursor CLTB 0.004 clathrin, light polypeptide isoform a CWC22 0.004 CWC22 spliceosome-associated protein homologHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
