
| Name: LOC100288291 | Sequence: fasta or formatted (145aa) | NCBI GI: 239746033 | |
|
Description: PREDICTED: hypothetical protein XP_002344174
|
Referenced in: RNA Polymerase and General Transcription Factors
| ||
|
Composition:
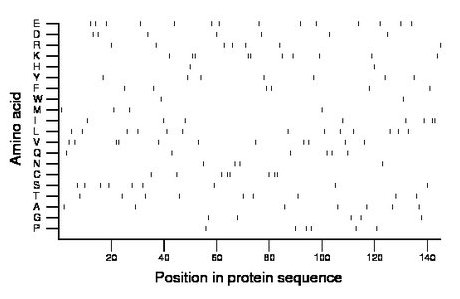
Amino acid Percentage Count Longest homopolymer A alanine 4.8 7 1 C cysteine 5.5 8 2 D aspartate 4.8 7 1 E glutamate 9.7 14 1 F phenylalanine 4.1 6 1 G glycine 3.4 5 1 H histidine 1.4 2 1 I isoleucine 5.5 8 2 K lysine 6.9 10 2 L leucine 9.0 13 1 M methionine 2.8 4 1 N asparagine 2.8 4 1 P proline 4.1 6 1 Q glutamine 4.1 6 1 R arginine 4.8 7 1 S serine 6.2 9 1 T threonine 6.2 9 1 V valine 7.6 11 2 W tryptophan 1.4 2 1 Y tyrosine 4.8 7 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002344174 LOC100291900 0.633 PREDICTED: similar to hCG2006635, partial LOC100290207 0.626 PREDICTED: hypothetical protein XP_002348308, parti... GTF2I 0.482 general transcription factor II, i isoform 3 GTF2I 0.482 general transcription factor II, i isoform 2 GTF2I 0.482 general transcription factor II, i isoform 1 GTF2I 0.482 general transcription factor II, i isoform 4 GTF2IRD2 0.446 GTF2I repeat domain containing 2 GTF2IRD2B 0.446 GTF2I repeat domain containing 2B GTF2IRD1 0.144 GTF2I repeat domain containing 1 isoform 1 GTF2IRD1 0.144 GTF2I repeat domain containing 1 isoform 2 LOC100132733 0.054 PREDICTED: similar to FLJ00310 protein LOC641765 0.047 PREDICTED: similar to hCG1983816 LOC641765 0.047 PREDICTED: similar to hCG1983816 LOC641765 0.047 PREDICTED: similar to hCG1983816 ZNF484 0.022 zinc finger protein 484 isoform b ZNF484 0.022 zinc finger protein 484 isoform a F7 0.014 coagulation factor VII isoform b precursor F7 0.014 coagulation factor VII isoform a precursor ZIM3 0.014 zinc finger, imprinted 3 ZNF337 0.014 zinc finger protein 337 ZNF334 0.011 zinc finger protein 334 isoform a ZNF334 0.011 zinc finger protein 334 isoform b CFC1 0.011 cryptic ZNF16 0.011 zinc finger protein 16 ZNF16 0.011 zinc finger protein 16 ZNF567 0.011 zinc finger protein 567 POLE 0.011 DNA-directed DNA polymerase epsilon LOC100287429 0.007 PREDICTED: hypothetical protein XP_002343926 isofor... LOC100287429 0.007 PREDICTED: hypothetical protein XP_002343925 isofor...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
