
| Name: LOC100131704 | Sequence: fasta or formatted (123aa) | NCBI GI: 169212142 | |
|
Description: PREDICTED: similar to NCOR
|
Referenced in:
| ||
|
Composition:
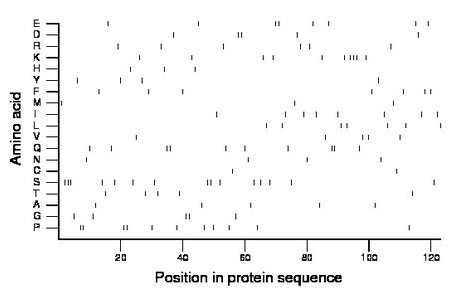
Amino acid Percentage Count Longest homopolymer A alanine 4.1 5 1 C cysteine 1.6 2 1 D aspartate 4.1 5 2 E glutamate 6.5 8 2 F phenylalanine 5.7 7 1 G glycine 4.1 5 2 H histidine 2.4 3 1 I isoleucine 6.5 8 1 K lysine 8.1 10 3 L leucine 5.7 7 1 M methionine 2.4 3 1 N asparagine 3.3 4 1 P proline 8.9 11 2 Q glutamine 8.1 10 2 R arginine 4.9 6 1 S serine 12.2 15 3 T threonine 4.1 5 1 V valine 4.1 5 1 W tryptophan 0.0 0 0 Y tyrosine 3.3 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to NCOR NCOR1 0.410 nuclear receptor co-repressor 1 NCOR2 0.145 nuclear receptor co-repressor 2 isoform 2 NCOR2 0.145 nuclear receptor co-repressor 2 isoform 1 EIF4B 0.031 eukaryotic translation initiation factor 4B LOC100133747 0.031 PREDICTED: hypothetical protein SH3BP5L 0.031 SH3-binding domain protein 5-like EWSR1 0.018 Ewing sarcoma breakpoint region 1 isoform 5 EWSR1 0.018 Ewing sarcoma breakpoint region 1 isoform 4 EWSR1 0.018 Ewing sarcoma breakpoint region 1 isoform 3 EWSR1 0.018 Ewing sarcoma breakpoint region 1 isoform 1 EWSR1 0.018 Ewing sarcoma breakpoint region 1 isoform 2 CECR2 0.018 cat eye syndrome chromosome region, candidate 2 [Ho... FAM21D 0.018 PREDICTED: hypothetical protein KIF20B 0.018 M-phase phosphoprotein 1 FAM21C 0.018 hypothetical protein LOC253725 TTF2 0.018 transcription termination factor, RNA polymerase II ... FAM21B 0.018 hypothetical protein LOC55747 QSK 0.013 serine/threonine-protein kinase QSK ZNF837 0.013 zinc finger protein 837 isoform 1 ZNF837 0.013 zinc finger protein 837 isoform 2 TTC23L 0.013 tetratricopeptide repeat domain 23-like FAM21A 0.013 hypothetical protein LOC387680 CCDC144B 0.013 coiled-coil domain containing 144B CCDC144A 0.013 coiled-coil domain containing 144A DRD2 0.013 dopamine receptor D2 isoform short DRD2 0.013 dopamine receptor D2 isoform long GRM7 0.013 glutamate receptor, metabotropic 7 isoform b precurs... GRM7 0.013 glutamate receptor, metabotropic 7 isoform a precurso... LOC100291914 0.013 PREDICTED: hypothetical protein XP_002345991Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
