
| Name: LOC100292268 | Sequence: fasta or formatted (164aa) | NCBI GI: 239755722 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
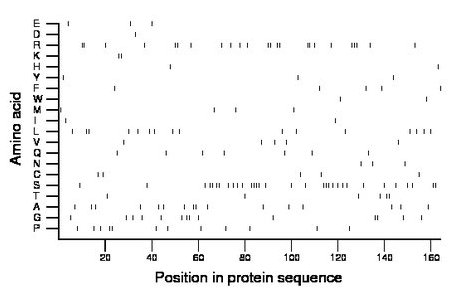
Amino acid Percentage Count Longest homopolymer A alanine 9.8 16 2 C cysteine 3.0 5 1 D aspartate 0.6 1 1 E glutamate 1.8 3 1 F phenylalanine 3.0 5 1 G glycine 8.5 14 2 H histidine 1.2 2 1 I isoleucine 1.2 2 1 K lysine 1.2 2 2 L leucine 9.8 16 2 M methionine 2.4 4 1 N asparagine 1.8 3 1 P proline 7.3 12 2 Q glutamine 4.3 7 1 R arginine 14.6 24 3 S serine 19.5 32 4 T threonine 3.7 6 2 V valine 3.0 5 1 W tryptophan 1.2 2 1 Y tyrosine 1.8 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100288159 1.000 PREDICTED: hypothetical protein LOC100288159 1.000 PREDICTED: hypothetical protein XP_002343179 PPRC1 0.063 peroxisome proliferator-activated receptor gamma, co... SCAF1 0.046 SR-related CTD-associated factor 1 C1orf63 0.040 hypothetical protein LOC57035 ZRANB2 0.036 zinc finger protein 265 isoform 1 ZRANB2 0.036 zinc finger protein 265 isoform 2 CHERP 0.036 calcium homeostasis endoplasmic reticulum protein [... RLIM 0.033 ring finger protein, LIM domain interacting RLIM 0.033 ring finger protein, LIM domain interacting HRH3 0.033 histamine receptor H3 SFRS16 0.033 splicing factor, arginine/serine-rich 16 LOC643446 0.030 PREDICTED: similar to ribonucleic acid binding prot... LOC643446 0.030 PREDICTED: similar to ribonucleic acid binding prot... TNRC18 0.030 trinucleotide repeat containing 18 RNPS1 0.030 RNA-binding protein S1, serine-rich domain RNPS1 0.030 RNA-binding protein S1, serine-rich domain RTN4 0.026 reticulon 4 isoform A LOC643446 0.026 PREDICTED: similar to ribonucleic acid binding prot... LOC643446 0.026 PREDICTED: similar to ribonucleic acid binding prot... LOC643446 0.026 PREDICTED: similar to ribonucleic acid binding prot... LOC643446 0.026 PREDICTED: similar to ribonucleic acid binding prote... SFRS2 0.026 splicing factor, arginine/serine-rich 2 FBXO41 0.026 F-box protein 41 SRRM2 0.026 splicing coactivator subunit SRm300 TTC24 0.023 tetratricopeptide repeat domain 24 TRIOBP 0.023 TRIO and F-actin binding protein isoform 6 C3orf71 0.023 hypothetical protein LOC646450 FLJ37078 0.023 hypothetical protein LOC222183Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
