
| Name: ACPT | Sequence: fasta or formatted (426aa) | NCBI GI: 14861860 | |
|
Description: testicular acid phosphatase precursor
|
Referenced in: Phosphatases and Sulfatases
| ||
|
Composition:
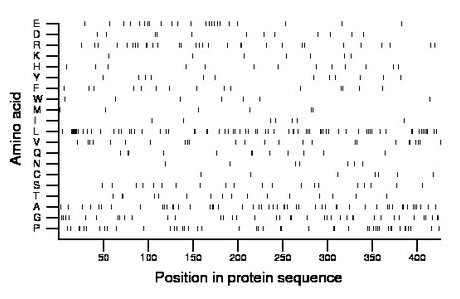
Amino acid Percentage Count Longest homopolymer A alanine 12.0 51 2 C cysteine 1.6 7 1 D aspartate 2.8 12 1 E glutamate 5.4 23 2 F phenylalanine 3.5 15 1 G glycine 9.2 39 2 H histidine 3.1 13 1 I isoleucine 1.6 7 1 K lysine 1.6 7 1 L leucine 16.7 71 7 M methionine 1.4 6 1 N asparagine 1.6 7 1 P proline 10.6 45 2 Q glutamine 2.8 12 2 R arginine 7.3 31 2 S serine 4.7 20 1 T threonine 3.8 16 2 V valine 5.9 25 2 W tryptophan 1.6 7 1 Y tyrosine 2.8 12 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 testicular acid phosphatase precursor ACP2 0.348 acid phosphatase 2, lysosomal isoform 1 precursor [Ho... ACPP 0.322 acid phosphatase, prostate long isoform precursor [... ACPP 0.322 acid phosphatase, prostate short isoform precursor [H... ACP2 0.146 acid phosphatase 2, lysosomal isoform 2 ACP6 0.086 acid phosphatase 6, lysophosphatidic ACPL2 0.046 acid phosphatase-like 2 ACPL2 0.046 acid phosphatase-like 2 LOC645781 0.014 PREDICTED: similar to mCG141871 LOC645781 0.014 PREDICTED: similar to mCG141871 LOC645781 0.014 PREDICTED: similar to mCG141871 MESP1 0.012 mesoderm posterior 1 PTPRN2 0.012 protein tyrosine phosphatase, receptor type, N poly... PTPRN2 0.012 protein tyrosine phosphatase, receptor type, N poly... PTPRN2 0.012 protein tyrosine phosphatase, receptor type, N poly... TNFRSF12A 0.009 tumor necrosis factor receptor superfamily, member 12... LOC392275 0.008 PREDICTED: similar to sphingomyelin phosphodiestera... LOC392275 0.008 PREDICTED: similar to sphingomyelin phosphodiestera... LOC100290470 0.007 PREDICTED: hypothetical protein XP_002347617 LOC100287301 0.007 PREDICTED: hypothetical protein XP_002343447 RELA 0.007 v-rel reticuloendotheliosis viral oncogene homolog ... RELA 0.007 v-rel reticuloendotheliosis viral oncogene homolog ... AXIN2 0.007 axin 2 TCHP 0.007 trichoplein TCHP 0.007 trichoplein INF2 0.007 inverted formin 2 isoform 2 INF2 0.007 inverted formin 2 isoform 1 PELP1 0.006 proline, glutamic acid and leucine rich protein 1 [... CCNK 0.006 cyclin K isoform 1 FLJ22184 0.006 PREDICTED: hypothetical protein FLJ22184Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
