
| Name: LOC100287396 | Sequence: fasta or formatted (398aa) | NCBI GI: 239744219 | |
|
Description: PREDICTED: hypothetical protein XP_002343076
| Not currently referenced in the text | ||
|
Composition:
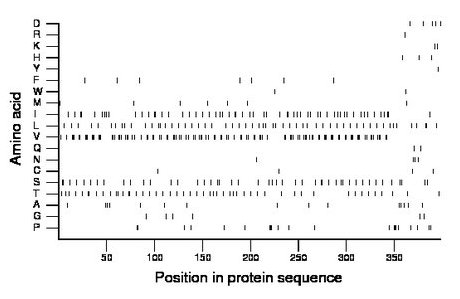
Amino acid Percentage Count Longest homopolymer A alanine 3.8 15 1 C cysteine 1.0 4 1 D aspartate 1.3 5 1 E glutamate 0.0 0 0 F phenylalanine 1.8 7 1 G glycine 1.5 6 1 H histidine 0.8 3 1 I isoleucine 14.6 58 2 K lysine 0.5 2 1 L leucine 14.1 56 2 M methionine 1.8 7 1 N asparagine 1.0 4 1 P proline 5.3 21 3 Q glutamine 0.5 2 1 R arginine 0.3 1 1 S serine 11.6 46 2 T threonine 12.6 50 2 V valine 27.1 108 3 W tryptophan 0.5 2 1 Y tyrosine 0.3 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002343076 LOC100289996 0.972 PREDICTED: hypothetical protein XP_002347212 LOC100293520 0.111 PREDICTED: hypothetical protein LOC100287207 0.111 PREDICTED: hypothetical protein XP_002342660 LOC100294237 0.111 PREDICTED: hypothetical protein LOC100294174 0.111 PREDICTED: hypothetical protein LOC100294124 0.111 PREDICTED: hypothetical protein LOC100294292 0.102 PREDICTED: hypothetical protein XP_002344066 LOC388849 0.096 PREDICTED: similar to hCG1732289 LOC100291351 0.095 PREDICTED: hypothetical protein XP_002347312 ABCA5 0.028 ATP-binding cassette, sub-family A , member 5 ABCA5 0.028 ATP-binding cassette, sub-family A , member 5 LOC100131552 0.027 PREDICTED: hypothetical protein LOC100131552 0.027 PREDICTED: hypothetical protein LOC100131552 0.027 PREDICTED: hypothetical protein OR4K2 0.025 olfactory receptor, family 4, subfamily K, member 2 ... LOC730474 0.025 PREDICTED: hypothetical protein LOC100294470 0.024 PREDICTED: hypothetical protein, partial GPR115 0.022 G-protein coupled receptor 115 LOC388849 0.019 PREDICTED: similar to hCG1732289 OR10J5 0.019 olfactory receptor, family 10, subfamily J, member 5... MUC16 0.018 mucin 16 OPRK1 0.018 opioid receptor, kappa 1 HAVCR1 0.016 hepatitis A virus cellular receptor 1 HAVCR1 0.016 hepatitis A virus cellular receptor 1 LOC100290205 0.016 PREDICTED: hypothetical protein XP_002348254 ABCA6 0.016 ATP-binding cassette, sub-family A, member 6 IFI27 0.016 interferon, alpha-inducible protein 27 isoform 2 [Ho... TAS2R46 0.016 taste receptor, type 2, member 46 ATP6 0.016 ATP synthase F0 subunit 6Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
