Domain Structure of Proteins
Many proteins can be readily parsed by sequence similarity into domains with specific functions. Other domains are defined by a relatively small number of noncontiguous, conserved residues, in some cases with variable spacing. Members of these families may be missed with searches using programs like BLASTP but detected with other pattern-matching methods.Multifunctional proteins
The human genome encodes a number of proteins with multiple enzymatic activities. These functions are sometimes encoded in separate proteins in other species. Examples are found in both pyrimidine and purine metabolism. An interesting case of the reverse is seen with enzymes in galactose utilization where the activities of separate human enzymes are found in a single yeast protein.Fatty acid synthase (FASN) is one of the most dramatic examples of multiple enzymatic activities encoded in a single polypeptide. Some of its domains have been found in separate mitochondrial proteins. Fatty acid oxidation also provides examples of proteins with separate and fused domains encoding different activities.
In some cases, domains with very different functions are found in a single protein. One interesting case is YARS, the cytoplasmic tyrosyl-tRNA synthetase. In addition to its familiar function in translation, it contains a domain also found in a protein that is a cytokine precursor.
Protein kinase domains are found in proteins that vary greatly in structure. An extreme example is the kinase domain in TTN (titin; also several other isoforms; see Muscle), the largest human protein. Although such domains are generally easy to locate by sequence similarity, largely unrelated sequences may have similar types of activities, as is observed with the RIO kinases.
Duplications
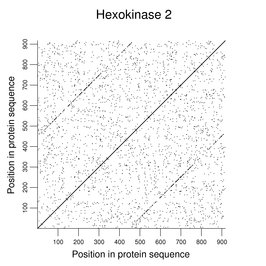
Some human enzymes have evolved by duplication of a smaller functional unit. Clear examples can be seen among the hexokinases. The following figure shows a dot plot of HK2 against itself (word size of 2) with the duplication easily seen.
A similar duplication is found in an isoform of the angiotensin-converting enzyme ACE (also isoform; see Cardiovascular System). Certain serine proteases have multiple protease domains.
One of the most interesting and complex examples of duplications involves the ubiquitin genes. This family includes genes encoding proteins with exact duplications of the ubiquitin protein sequence, fusions to other genes that encode identical protein sequences in their ubiquitin domains, and additional family members with related domains.
The ATP-binding cassette family includes half transporters that require dimerization for function and larger proteins that contain an internal duplicated region.
Duplicated regions are seen in many other types of proteins. One interesting case is the terminal duplication in pleckstrin (see Inositol Pathways). MUC16 (see Mucins) contains a large tandem array of a sequence not found in other proteins. The sequence of EML5 (see Tubulin and Microtubules) is essentially a triplication of another family member.
Regulatory domains
Many protein domains have regulatory functions. Sometimes these domains are found in otherwise unrelated proteins or separately as small proteins.The human genome contains multiple calmodulin genes that encode identical proteins. In addition to calmodulin, many other proteins have calmodulin-related domains. These domains are found in different parts of proteins (see Calmodulin and Calcium).
An interesting case is the sterol-binding regions in proteins with diverse functions (see Cholesterol Biosynthesis). Related sequences are present in an enzyme, a chaperone, a cholesterol trafficking protein, and others.
Structural domains
Many domains involved in mediating protein interactions are described in the chapter devoted to that topic and its section on how they are defined (see Domains, Motifs, and Composition). These structural domains are generally found only in eukaryotic species. Many, such as WD repeats, are found in diverse eukaryotes. Some have more limited distribution; for example, PDZ domains are not readily identified by sequence similarity in yeast. Often, these units are named for the proteins where they were first recognized. Frequently, they are present in multiple copies and in complex combinations. Proteins with such domains include immunoglobulins, epidermal growth factor, and proteins with fibronectin-type repeats. Many of the proteins involved in the growth and development of neurons contain these and other domains.A number of functional domains were first noted in the SRC (also alt mRNA) tyrosine kinase. The SH2 and SH3 domains are found in many proteins.
Some protein domains are associated with modifications to specific residues of the proteins. One example is the vitamin-K-dependent modification of glutamates in coagulation factors and related proteins.
Although a protein domain may be generally associated with a family of proteins with related functions, it also may be found in proteins with very different functions. One such example is the hemopexin-related proteins.
Notes and references
Many references and other information for individual genes can be found in the RefSeq entries linked via the pages for the proteins mentioned in this section. A table of these entries (with the corresponding gene identifiers) and a collection of their sequences also are available.TTN has alternate products that can be accessed via the link in this section.
See also the additional reading for this chapter.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.